Wheat is prone to a swarm of different pests and diseases. Given its importance, this is unsurprising; the crop is the most widely grown in the world, and provides 18% of humankind’s calories[1]FAOSTAT. Pests and pathogens take a large bite out of wheat yields – adding up to 21% of the world’s projected harvest[2]Savary, S. et al. The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 3, 430–439 (2019)., or 209 million tonnes. Recapturing some of this yield can only be a good thing for food security, but how do we get there? We believe a wheat R gene atlas could help to tackle this problem.
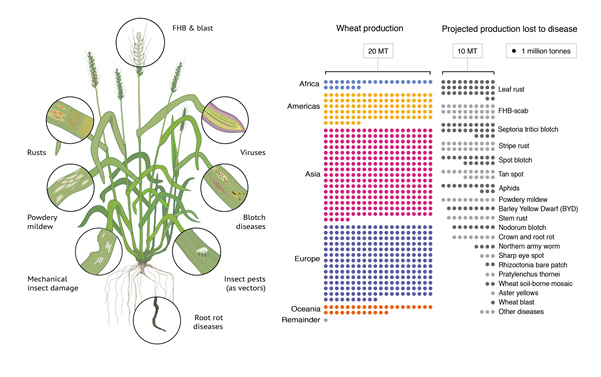
The symptoms (left) and production losses (right) associated with the major pests and pathogens of wheat, based on data from Savary et al. 2019. This figure was reproduced from Hafeez et al. 2021.
Currently, European wheat is heavily treated with fungicides, but chemical controls are not always available to growers due to lack of funds or heavy regulation, the latter of which is an increasing problem in the EU. It is imperative that we safeguard our daily bread, whilst also growing crops more sustainably. We can do this by enhancing host resistance, and by ensuring that sources of host resistance are managed appropriately so they provide effective control of pests and pathogens in the long term.
A wheat R gene atlas could serve as a solution. Host resistance (R) genes work by recognising corresponding molecules in the pathogen – called effectors. By monitoring the effectors present in pathogen or pest populations, combinations of R genes (known as stacks) that most efficiently control disease threats could be developed.

An ‘Atlas’ to support wheat breeding.
In our review, ‘Creation and judicious application of a wheat resistance gene atlas’, we detail how this could be done at a scale large enough to control wheat’s major diseases. Whole-genome sequencing could be carried out on diversity panels of wheat, its progenitors and some wild and domesticated relatives (such as rye). Then, association genetics could be used to look for correlations between the host genotype and disease resistance or susceptibility – and the genes responsible for these traits could be identified.
How could we apply the wheat R gene atlas to breeding?
When pathogens are repeatedly exposed to host resistance genes, then virulence is more likely to be selected for – especially when R genes are working alone. “The life of ‘big’ winter wheat varieties in West European countries has become shorter and shorter, and is currently about 3 to 5 years. Replacement of varieties is fast,” Viktor Korzun at KWS SAAT SE & Co. KGaA, Germany, told us – one of the many breeders we spoke to when developing the ideas in our review.
We could break this boom-and-bust cycle by aiming for resistance that is difficult to break down and so is effective long-term (durable). Combined with effective monitoring of pest and pathogen populations, an R gene atlas could be used to identify combinations of R genes that would provide broadly effective and durable resistance against major threats.
One problem is that it can take many years to introduce new genes, especially from wild backgrounds. To control just three major diseases effectively, a minimum of 12 broad-spectrum genes would be required. Even when using techniques such as speed breeding[3]Watson, A. et al. Speed breeding is a powerful tool to accelerate crop research and breeding. Nat. Plants 4, 23–29 (2018). to reduce generation times, it would take up to four years to develop a cultivar with so many R genes through conventional breeding. After all of this, the cultivar will have fallen behind in yield – and all 12 genes would need to be bred into a new background. Alternatively, a GM stack of R genes could be directly introduced into a desired background. This has recently been demonstrated in wheat with a 5-transgene stack[4]Luo, M. et al. A five-transgene cassette confers broad-spectrum resistance to a fungal rust pathogen in wheat. Nat. Biotechnol. (2021). doi:10.1038/s41587-020-00770-x which confers highly effective resistance to stem rust – a huge leap towards accelerating wheat breeding.
Another obstacle is understanding the history of particular R genes – knowing which genes are already available to breeders and how they have been exposed to pests and pathogens. We spoke to Wang Xiue at Nanjing Agricultural University in China, who said that in China it is necessary to narrow down “the gap between research and breeding. The knowledge about which gene combinations are durable should be transferred to the breeders for developing varieties with durable resistance. Currently, if a variety has good enough resistance to dominant diseases, farmers will grow it.” We also spoke to Sridhar Bhavani, the head of rust research at CIMMYT, who told us: “an atlas of R genes would be a valuable resource to better understand the genetics and to dissect the complex resistance present in CIMMYT germplasm. Pedigrees are humongous, and it can be hard to figure out at times the contributions of certain genotypes and their interactions in enhancing resistance in different genetic backgrounds.”
Best Practices
Good gene stewardship practices, described in detail by Robert Bowden, USDA-ARS in our review, involve “the careful and responsible management of R genes to remain effective during prolonged use”. Some “inherently durable” genes (such as the famous Sr2) may not require stewardship, but race-specific genes generally do – so it is important that we find some way to monitor and control their release. Another aspect of gene stewardship would be the safeguarding of genes that are new to farmers’ fields, such as those from wild species, through patenting. Importantly, having perfect markers available for R genes would allow breeders to ascertain the R gene complements of their lines. This could enable reward schemes by release committees to incentivise breeding for cultivars with many and varied sources of disease resistance. Filippo Bassi at ICARDA and Hannah Robinson at Intergrain, Australia, said that working with release committees could be a viable strategy in their regions; on the other hand, Zakkie Pretorius and Willem Boshoff at the University of the Free State, South Africa, felt that “since the focus is currently on higher yielding varieties, efforts to enforce gene stewardship at release meetings will not be acceptable to all breeders and producers in South Africa.” More work needs to be done to develop effective strategies to encourage both gene stewardship and breeding for durable resistance.
We heard a similar tale of breeding priorities from many of those we spoke to – as Brian Steffenson, based at the University of Minnesota, put it: “what dictates success in the marketplace is yield: bushels, bushels, bushels.” However, if R gene stacks can remove the threats from major diseases, breeding can continue to focus on yield and other traits – so everyone would win, hopefully on a global scale.
Getting ready for the future
Overall, the wheat R gene atlas would benefit breeders by making more information available to them. There could also be benefits for other related grass crops, such as barley and rye. More broadly, amassing so much information about the genetic basis of so many diseases would help to understand related pathosystems in other crops, and plant immunity in general. Looking towards the future, Korzun told us: “the requirement for broad, durable resistance will undoubtably increase – we, as plant scientists and breeders, need to be ready.”
References
↑1 FAOSTAT.
↑2 Savary, S. et al. The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 3, 430–439 (2019).
↑3 Watson, A. et al. Speed breeding is a powerful tool to accelerate crop research and breeding. Nat. Plants 4, 23–29 (2018).
↑4 Luo, M. et al. A five-transgene cassette confers broad-spectrum resistance to a fungal rust pathogen in wheat. Nat. Biotechnol. (2021). doi:10.1038/s41587-020-00770-x
References
| ↑1 | FAOSTAT. |
| ↑2 | Savary, S. et al. The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 3, 430–439 (2019). |
| ↑3 | Watson, A. et al. Speed breeding is a powerful tool to accelerate crop research and breeding. Nat. Plants 4, 23–29 (2018). |
| ↑4 | Luo, M. et al. A five-transgene cassette confers broad-spectrum resistance to a fungal rust pathogen in wheat. Nat. Biotechnol. (2021). doi:10.1038/s41587-020-00770-x |