Plant diseases remain a constant scourge to all agricultural crop production. Despite intensive efforts and large expenditure to control diseases using genetic resistance, fungicides and agronomic practices like crop rotation, diseases still annually account for a 10-15% average yield loss in all crops. Of course, in particularly bad seasons entire crops can be lost in local areas causing regional economic hardship and in some instances malnutrition. Genetic resistance is highly advantageous given that it is inexpensive, doesn’t rely on chemicals and can be readily deployed in all crop growing areas using existing infrastructure.
Why doesn’t plant genetic resistance last?
Genetic resistance is often only transient due to the constant evolution of disease-causing pathogens. Faced with these ever-changing pathogen populations breeders are forced to constantly identifying new resistance genes and introducing them into elite cultivars to maintain crop protection. Resistance genes have been cloned from many plant species and most resistance gene products recognises a corresponding molecule produced by a pathogen. Upon this recognition a defense response is activated in the plant to give resistance. However, these pathogen molecules are highly variable and often dispensable. Pathogen populations can rapidly change or lose a recognised molecule to avoid recognition by a plant resistance gene product. Therefore, the resistance conferred by this resistance gene breaks down.
How to overcome pathogen evolution?
Our group is interested in the resistance of wheat to wheat rust diseases caused by the fungal pathogens Puccinia graminis f.sp. tritici (wheat stem rust), P. striiformis f.sp. tritici (wheat stripe rust and P. triticina (wheat leaf rust). These three fungal diseases are a major disease threat to all wheat growing regions in the world. An ever-growing number of wheat rust resistance genes have been cloned with most of these gene products each recognising a unique molecule produced by the rust pathogen. If these genes are used singularly they tend to breakdown rapidly due to the dispensable nature of the pathogen molecules being recognised.
However, while the pathogen can readily lose one recognised molecule, it becomes progressively harder and harder to lose more and more of these molecules all at the same time. This is why combining resistance genes together gives more stable resistance. It is like combining locks on a door, the more you use the harder it is to break in and you have to overcome every lock at the same time to get in. Resistance genes to wheat rusts, and other diseases, can be combined by breeding but this is laborious to do and even harder to keep all the genes together in subsequent breeding as each gene is usually located in a different region of the genome.
Genestacks
To overcome these problems, we have developed a gene stack[1]Luo M, Xie L, Chakraborty S, Wang A, Matny O, Jugovich M, Kolmer JA, Richardson T, Bhatt D, Hoque M, Patpour M, Sørensen C, Ortiz D, Dodds P, Steuernagel B, Wulff BBH, Upadhyaya NM, Mago R, … Continue reading.
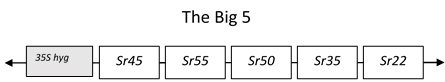
In this case we have used wheat stem rust resistance genes and physically combined 5 stem rust resistance genes into a single molecule, in addition to a 35S-hygromycin selectable marker gene[2]Richardson T, Thistleton J, Higgins TJ, Howitt C, Ayliffe M (2014) Efficient Agrobacterium transformation of elite wheat germplasm without selection. Plant Cell, Tissue and Organ Culture 119: 647- 659. This DNA construct we named the Big5 and then introduced all these genes into a single location in the wheat genome using a bacterium (Agrobacterium) that naturally introduces DNA into plant cells. As these genes are physically linked they will remain together as a single unit when used for breeding and the polygenic nature of the resistance promises to increase the longevity of all the resistance genes in the stack. This was challenging because combining all these large genes together results in a very large molecule to manipulate and transform. To achieve this, we had to develop a unique sequential cloning system and screen a large number of transgenic wheat lines to find ones that had all the genes present. These lines, however, showed very high levels of field resistance to wheat stem rust and were resistant to a diverse global panel of wheat stem rust isolates.

A global problem requires a global collaboration
For this research to occur we fortunately had financial support from the visionary 2Blades Foundation and had world leading international collaborators help from the University of Minnesota (USA), Aarhus University (Denmark), John Innes Centre (UK), United States Department of Agriculture and Xianjing University (China). The global nature of wheat rust diseases requires these global collaborations to combat them.
Future gene stacks
We are currently developing more wheat rust resistance gene stacks, now using both new recently cloned stem rust resistance genes and stripe rust resistance genes. We are also combining gene stacks to develop plants containing even more functional rust resistance genes (i.e. 10 genes at two loci) in an elite African cultivar, Robin. This material will be field trialled in Africa given the recurrent emergence of new wheat stem rusts from Africa including the Ug99 and Digalu lineages. The gene stacking approach we have developed is not just limited to rust resistance genes and wheat of course and is applicable to any plant species for which resistance genes have been cloned and a transformation system is available. Most notably a three resistance gene stack to potato late blight has been developed[3]Ghislain M, Byarugaba AA, Magembe E, Njoroge A, Rivera C, Roman ML, Tovar JC, Gamboa S, Forbes GA, Kreuse JF, Barekye A, KIggundu A (2018) Stacking three late blight resistance genes from wild … Continue reading. This approach therefore has broad potential application to many agricultural and horticultural plant species and offers a way to increase resistance durability.
References
↑1 Luo M, Xie L, Chakraborty S, Wang A, Matny O, Jugovich M, Kolmer JA, Richardson T, Bhatt D, Hoque M, Patpour M, Sørensen C, Ortiz D, Dodds P, Steuernagel B, Wulff BBH, Upadhyaya NM, Mago R, Periyannan S, Lagudah E, Freedman R, Reuber TL, Steffenson BJ, Ayliffe M (2021) A five-transgene cassette confers broad-spectrum resistance to a fungal rust pathogen in wheat. Nature Biotechnology DOI: 10.1038/s41587-020-00770-x.
↑2 Richardson T, Thistleton J, Higgins TJ, Howitt C, Ayliffe M (2014) Efficient Agrobacterium transformation of elite wheat germplasm without selection. Plant Cell, Tissue and Organ Culture 119: 647- 659
↑3 Ghislain M, Byarugaba AA, Magembe E, Njoroge A, Rivera C, Roman ML, Tovar JC, Gamboa S, Forbes GA, Kreuse JF, Barekye A, KIggundu A (2018) Stacking three late blight resistance genes from wild species directly into African highland potato varieties confers complete field resistance to local blight races. Plant Biotechnology Journal 17: 1119-1129.
References
| ↑1 | Luo M, Xie L, Chakraborty S, Wang A, Matny O, Jugovich M, Kolmer JA, Richardson T, Bhatt D, Hoque M, Patpour M, Sørensen C, Ortiz D, Dodds P, Steuernagel B, Wulff BBH, Upadhyaya NM, Mago R, Periyannan S, Lagudah E, Freedman R, Reuber TL, Steffenson BJ, Ayliffe M (2021) A five-transgene cassette confers broad-spectrum resistance to a fungal rust pathogen in wheat. Nature Biotechnology DOI: 10.1038/s41587-020-00770-x. |
| ↑2 | Richardson T, Thistleton J, Higgins TJ, Howitt C, Ayliffe M (2014) Efficient Agrobacterium transformation of elite wheat germplasm without selection. Plant Cell, Tissue and Organ Culture 119: 647- 659 |
| ↑3 | Ghislain M, Byarugaba AA, Magembe E, Njoroge A, Rivera C, Roman ML, Tovar JC, Gamboa S, Forbes GA, Kreuse JF, Barekye A, KIggundu A (2018) Stacking three late blight resistance genes from wild species directly into African highland potato varieties confers complete field resistance to local blight races. Plant Biotechnology Journal 17: 1119-1129. |